In this article, we present a monitoring concept for arthropods that uses limited financial and human resources to offer a good compromise between a few individual, isolated traps and a complete survey.
The Entomological Association of Krefeld found in 2017 that the biomass of flying insects in nature reserves had declined by 75 % within 27 years. Since that time at the latest, the general public has been aware of the issue of insect mortality, and it has been the subject of sometimes heated debate. Subsequently, there has, among other things, been a petition for a referendum on “Save the Bees”, and corresponding changes have been made to the law.
In specialist circles, there are increasing indications of a dramatic decline in the populations that has been taking place for some time. There have however been no corresponding long-term studies so far. It will probably be some time before these are available on a sufficient scale - time that we do not really have, given the rapid progression of environmental changes. Overall, the need for short-term information on biodiversity has increased enormously in recent years. In a densely forested country like Germany, this naturally also affects the forest habitat.
Limits of classical taxonomy
Classical taxonomy alone can no longer cope with these challenges. This is due on the one hand to the increased number of samples and on the other hand to the time-consuming determination of species. What is also the case is that experienced taxonomists for the identification of many species groups, such as arthropods, which include numerous insect species, are themselves almost a “dying species” today. The few remaining taxonomists sometimes already have work to keep them busy for years to come.
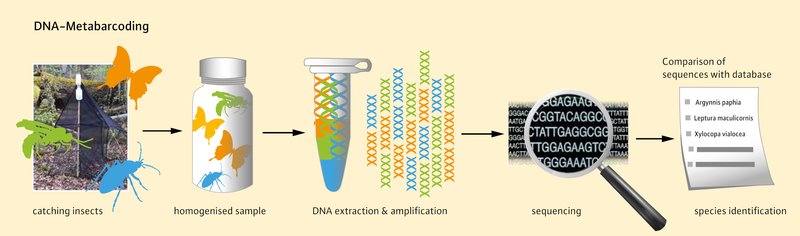
Today, new methods such as DNA metabarcoding are available to fill this gap. This method can be used to identify individual species from a mixed sample that may contain several thousand individuals. The advantage over the classic method is that a result is achieved quickly at a relatively manageable cost. However, DNA metabar coding also has some disadvantages. For example, although this method can very sensitively determine which genetic hereditary information was contained in the sample, it does not currently provide any abundances or frequencies of occurrence. In addition, the entire mixed sample must first be homogenised, i.e. pulverised, so that the individual specimens are subsequently no longer available for further investigations. This therefore does not replace classical taxonomy, but merely complements it.
How does DNA metabarcoding work?
The captured arthropods, e.g. from a flight interception trap, are first dried and then weighed. These working steps are important for later ecological analyses (dry biomass). Optionally, the mixed sample of arthropods can then be separated by size into two or more fractions (size fractionation). This is always useful when species of different sizes (very small and very large individuals) are present in a sample. There is a danger here that large specimens such as bumblebees or large beetles with a lot of DNA “cover up” the smaller individuals (which have less DNA), so that they are then under-represented. Only after size fractionation is the sample then homogenised and the DNA extracted.
The genetic marker used for the analysis is subunit I of cytochrome C oxidase (COI). This enzyme is part of the respiratory chain and plays a central role in the energy metabolism of many living organisms. Our genetic marker COI is not located like most genes in the cell nucleus, but in the so-called mitochondria. These are often referred to as the power plants of the cells and have their own genetic material, which is usually only passed on by the females. This and the fact that the DNA sequence of COI shows more differences between different species than other mitochondrial genes makes it particularly interesting for taxonomic purposes.
For further analysis, the COIs are specifically amplified, after which the respective individual sequences of DNA components (bases) of the animals can be determined using a next-generation sequencing procedure. The read DNA sequences of the COIs differ more or less depending on the species, and can then be compared with reference sequences in databases. If a species is already known from previous metabarcoding studies, it can be assigned to the corresponding sequence. If this is not the case, an assignment can still be made years later if the species concerned is then available in the comparative databases.
In principle, however, the DNA sequences determined alone, without comparison sequences, provide indications of species diversity and a measure of biodiversity, since samples whose sequences show a difference of more than 3 % are usually evaluated as different species. For ecological questions on biodiversity in which it is not of primary importance to know the individual species by name, these data are therefore very valuable.
Application in the Forest Climate Fund project “natWald100”
As part of the “natWald100” project funded by the Forest Climate Fund, the Bavarian State Institute of Forestry (LWF) and the Northwest German Forest Research Institute (NW-FVA) are investigating the effect of natural forest development on carbon storage and biodiversity in one hundred strict forest reserves across Germany. The LWF is working on the topic of biodiversity, including the biodiversity of arthropods. In each strict forest reserve, a combination of malaise, ground and flight interception traps was installed for this purpose, as all three trap types cover a very different spectrum of species. While malaise traps mainly catch positively phototactic, phytophagous, i.e. light-attracted, herbivorous insects such as flies (Diptera) and Hymenoptera (sawflies, wasps, ants, bees), flight interception traps tend to catch negatively phototactic insects such as deadwood beetles. Ground traps cover the ground fauna, which includes ground beetles (Carabidae), spiders and isopods.
All traps work with ethanol (80 %) as a preservative to secure the catch for later DNA analysis. The traps are emptied fortnightly from the beginning of May to the end of July. Altogether, a total of 1,800 individual samples are collected, each of which can contain several hundred or up to a thousand individuals. Due to the abundance of sample material, the current project would not be feasible within a reasonable time and financial framework without the use of DNA metabar coding.
Summary
For many scientific studies and monitoring approaches in the field of biodiversity, DNA metabarcoding offers a means of obtaining solid data on the diversity and occurrence of individual species in a timely manner and within a manageable budget. As a disadvantage, it must be accepted that no statements on abundances can be derived with this method and that the sample is lost during the analysis. However, the advantages of this method usually outweigh the disadvantages. Often, DNA metabar coding makes possible research projects that would not be feasible due to the short-term nature of the data requirements.